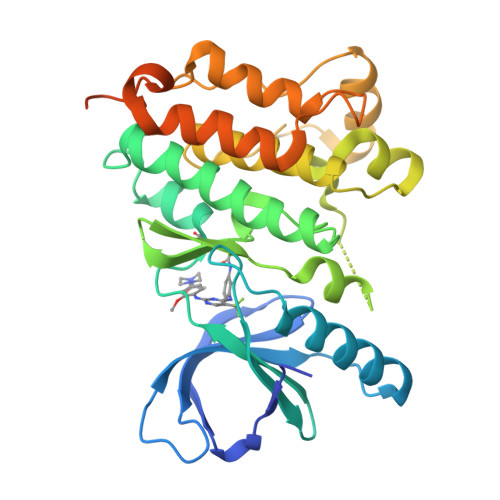
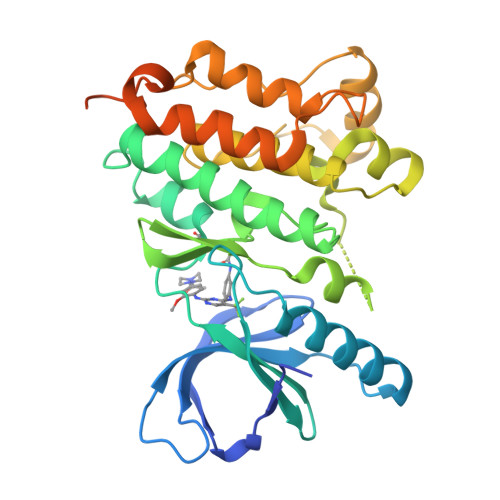
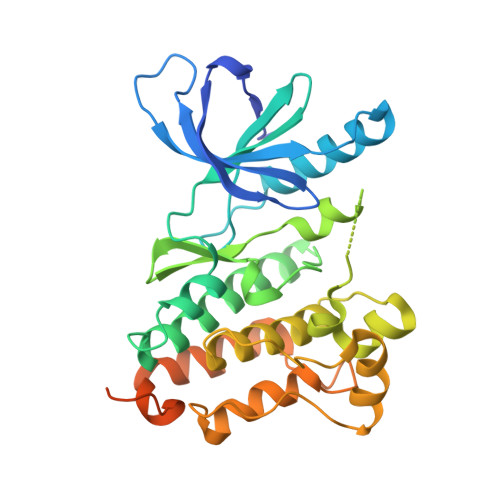
Proteome-wide Map of Targets of T790M-EGFR-Directed Covalent Inhibitors.
Niessen, S., Dix, M.M., Barbas, S., Potter, Z.E., Lu, S., Brodsky, O., Planken, S., Behenna, D., Almaden, C., Gajiwala, K.S., Ryan, K., Ferre, R., Lazear, M.R., Hayward, M.M., Kath, J.C., Cravatt, B.F.(2017) Cell Chem Biol 24: 1388-1400.e7
- PubMed: 28965727
- DOI: https://doi.org/10.1016/j.chembiol.2017.08.017
- Primary Citation of Related Structures:
5UWD - PubMed Abstract:
Patients with non-small cell lung cancers that have kinase-activating epidermal growth factor receptor (EGFR) mutations are highly responsive to first- and second-generation EGFR inhibitors. However, these patients often relapse due to a secondary, drug-resistant mutation in EGFR whereby the gatekeeper threonine is converted to methionine (T790M). Several third-generation EGFR inhibitors have been developed that irreversibly inactivate T790M-EGFR while sparing wild-type EGFR, thus reducing epithelium-based toxicities. Using chemical proteomics, we show here that individual T790M-EGFR inhibitors exhibit strikingly distinct off-target profiles in human cells. The FDA-approved drug osimertinib (AZD9291), in particular, was found to covalently modify cathepsins in cell and animal models, which correlated with lysosomal accumulation of the drug. Our findings thus show how chemical proteomics can be used to differentiate covalent kinase inhibitors based on global selectivity profiles in living systems and identify specific off-targets of these inhibitors that may affect drug activity and safety.
Organizational Affiliation:
Worldwide Medicinal Chemistry, La Jolla Laboratories, Pfizer Worldwide Research and Development, 10770 Science Center Drive, San Diego, CA 92121, USA; Oncology RU, La Jolla Laboratories, Pfizer Worldwide Research and Development, 10770 Science Center Drive, San Diego, CA 92121, USA. Electronic address: sherry.niessen@pfizer.com.